EDH 7916: Contemporary Research in Higher Education
Spring 2020
A course in quantitative research workflow for students in the higher education administration program at the University of Florida
Overview Course information Meeting location Getting started Schedule Lessons Assignments Questions About
Introduction
A system of Version Control: Introduction to Git / GitHub
From the git website http://git-scm.com/
Git is a free and open source distributed version control system designed to handle everything from small to very large projects with speed and efficiency.
Why use git?
With so many other (read: easier!) ways to share and collaborate on documents, why use git? Isn’t it a bit overkill to learn an entirely new syntax? Why not just email files or use something like DropBox? Because it is very easy to end up with something like this:
Credit: Jorge Chan
As complexity increases, so does the need for git
\[ Project = f(size, scope, collaborators) \]
As any part of that function grows, so too does the need for a work flow that:
- Allows for many moving parts
- Allows for peaceful collaboration (no overwrites)
- Allows for timely collaboration (synchronous)
- Allows for distant collaboration (centralized file location)
- Allows for version control (so you can go back if necessary)
Git was designed to do all these things, so it works better than other systems.
What’s the difference between git and GitHub?
Yep, you guessed it. Git is the system/language that supports version control. GitHub is an online service that will host your remote repositories, for free if public or a small fee if private. (Students get an education discount and some free repos. Check out https://education.github.com/.)
RStudio is nice because it provides an nice point-and-click interface for git. (Sourcetree and GitHub Desktop are also really nice GUIs.) If you want to run git at the command line, go for it! But using RStudio or another GUI is fine.
How does git/GitHub work?
Some notes on work flow (good habits)
- Always
pullfrom your local repo with your remote repo before starting any work. This makes sure you have the latest files. RStudio has apullbutton in the Git tab. - Don’t wait to
pushyour commits. Just like you save your papers every few lines or paragraphs, you should push to your remote repo. This way, you’re less likely to lose work in the event of a computer crash. Also, should you want to return to a prior version, small changes make it easier to find what you want. - Add useful
commitmessages. RStudio will make you add something. Don’t say “stuff.” A version history of 95 “stuff”s is pretty non-helpful. Also, I would keep it clean. Even in a private repository, work as if someone will see what you write someday. - Don’t freak out! If you accidentally push and overwrite or delete something on the remote, you can get it back. That’s the point of version control! Sometimes weird things like merges happen (two files with the same name are shoved together). They can be a pain to fix, but they can be fixed.
- Avoid tracking overly large files and pushing them to your remote repository. GitHub has a file size limit of 100 MB per file and 1 GB per repo. The history of large files compounds quickly, so think carefully before adding them. Usually this means that small datasets are okay; big ones are better backed up somewhere else and left on your machine.
- Remember: even in a private repository, your files are potentially
viewable. If any of your datasets or files are restricted, do no
push them remotely. Use
.gitignoreto make sure that git doesn’t track or push them.
Every class and work session should go like this:
pullfrom GitHub remote- do work
addorstage(if new file)commitwith messagepushback to GitHub remote
If you’d like you can also use terminal commands to accomplish the same things. Many people (including me) like to use the terminal commands directly.
- Pull:
git pull - Stage:
git add <filenames> - Committ:
git commit -m "<your message here>" - Push:
git push
That said, you should use whatever works best for you.
By far the most useful guide to working with R and git/GitHub is Jenny Bryan’s guide.
Some more notes on using Git: plain text
Git works best with plain text files. This is because it notes the differences across two plain text files rather than just copying and re-copying the same file over and over again as it changes. When you’ve only changed one word, git’s method of version control is much more efficient than making a whole new copy.
It’s also useful when you need to merge two files. If file B is just file A with new section, file B can be easily merged into file A by inserting the new section — just like you would add a paragraph in a paper you’re writing. R scripts are plain text. Some data files, like *.csv, are plain text. This is why git works really well with data analysis workflows.
On the other hand, git does not work as well with binary files. These files are stored in a format closer to what your computer understands, which comes with benefits. Data files, like Stata’s .dta and R’s *.Rdata, as well as MS Office files — *.docx, *.xlsx, *.pptx, *etc — are binary files. Git will keep track of your MS Word document, but due to its underlying structure, you won’t be able to merge and every small change will just make a whole new copy of the file. This is why we generally don’t commit large binary data files to Git: your repo just becomes larger and larger with each small change to your data.
Not-so-quick exercise
In this first exercise, we’ll practice using RStudio to pull down your student repository from GitHub. We’ll also practice making a few quick file changes, staging, commiting, and pushing them back to the remote.
A Language of Data Analysis: Introduction to R
R is a port of the S language, which was developed at Bell Labs. As a GNU project, R is open source and free to use and distribute. It can be installed and used on most major operating systems.
R is best thought of as an integrated language and environment that was designed with statistical computing and data analysis in mind. To that end, its structure is a balance between powerful mathematical computation and high-level functionality that can be used interactively (unlike compiled code). In other words, it’s a great tool for quantitative data analysis since it both allows you to investigate your data easily and, when the time comes, write robust programs.
Originally, R was probably best known for its graphing capabilities. As it has matured, it has grown in popularity among data scientists, who have increasingly extended its functionality through user-contributed packages. We will use a number of packages during this course.
RStudio: an integrated development environment (IDE) for R
To work with the R language, it helps to have an application. While R ships with one (you may see it on your computer as R.app), it’s pretty plain. RStudio, on the other hand, is a powerful IDE that does most everything R-related very well and with little fuss: run commands, write scripts, view output, interact with other languages and remote site, etc. There are other options for working with R, but RStudio is a great all-around program that we will use in this course.
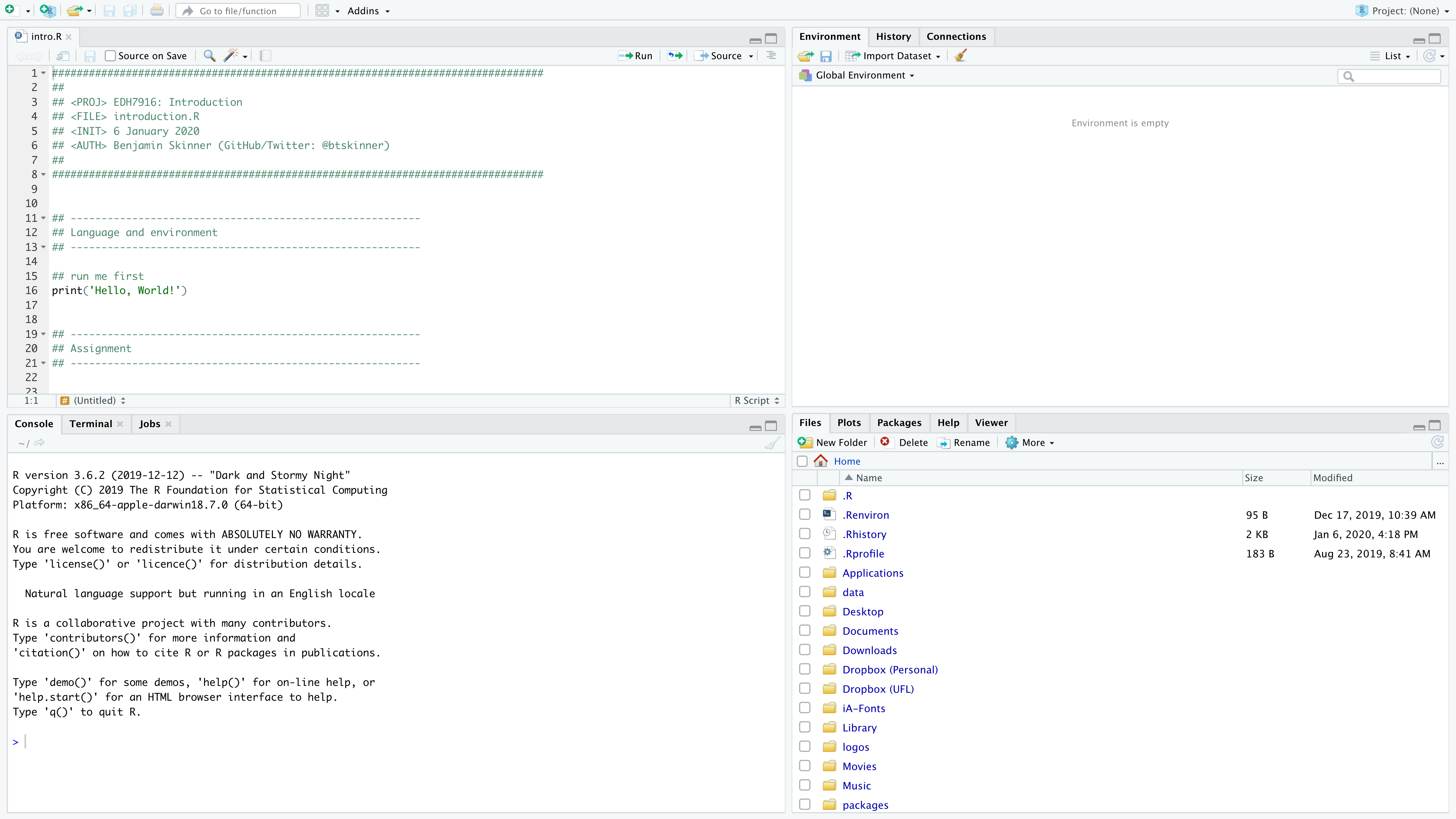
RStudio has 3-4 main frames:
- Console
- Script window (will be closed at first if you don’t have any scripts open)
- Environment / History / Connections
- Files / Plots / Packages / Help / Viewer
Each has a useful purpose, but for today, we’ll mostly focus on the console itself.
Quick exercise
If you haven’t already, try entering an equation in the console (like
1 + 1). Next, open the script associated with this module and run the first line. Welcome to R!
Assignment
R is a type of object-oriented programming environment. This means that R thinks of things in its world as objects, which are like virtual boxes in which we can put things: data, functions, and even other objects.
Before discussing data types and structures, the first lesson in R is
how to assign values to objects. In R (for quirky
reasons),
the primary means of assignment is the arrow, <-, which is a less than
symbol, <, followed by a hyphen, -.
## assign value to object x using <-
x <- 1
NOTE: You can also use a single equals sign, =, to assign a value
to an object: x = 1. Keep in mind, however, that since = sometimes
has other meanings in R and can be confused with ==, which is
different, it’s generally clearer to use <-.
But’s where’s the output?
R does exactly what you ask it to do — no more, no less. If you don’t ask it to return something, either explicitly from a function or implicitly by printing to the console, it won’t. This can be huge source of frustration to new users.
The good-ish news is that by default, R will print an object’s contents to the console if it’s the only thing you type in. Many functions similarly print to the console if you don’t assign the output to an object. You can see this when simply type a number or character into the console.
## when you input a number or character, R returns it back to you
1
[1] 1
"a"
[1] "a"
Basically, you’ve just told R “Here’s a 1” and R said “The content of
1 is 1”. Same for "a". The initial number in the square brackets
([1]) is telling you the index (place within the object) of the first
item. Since we only have one item, it’s just [1].
When you store something in an object, you can type the object’s name into the console to see what’s in it.
## what's in x?
x
[1] 1
A neat trick if you want to both assign a value and see the results
printed to the output is to to wrap the entire line in ().
## wrap in () to print after assignment
(x <- 5)
[1] 5
Quick exercise
Using the arrow, assign the output of
1 + 1tox. Next subtract 1 fromxand reassign the result tox. Show the value inx.
A NOTE ON “GOOD-ISH” Keep in mind how much data your object has /
might have when printing it to the console. For a small amount, printing
is just fine. But if you have, for example, a matrix with 1,000 columns
and 1 million rows, printing might not be a useful exercise. There are
other ways, such as the function head() that might be more useful in
these situations.
Comments
You may have noticed already, but comments in R are set off using the
hash or pound character at the beginning of the line: #. The comment
character tells R to ignore the line, that is, do not try to interpret
it as code you the user want run.
Quick exercise
Type the phrase “This is a comment” directly into the R console both with and without a leading “#”. What happens each time?
You may notice that I use two hashes. This is a stylistic tick that has
more to do with the editor I use than an R requirement. You can use only
a single # for your comments if you like.
Data types and structures
R uses variety of data types and structures to represent and work with data. There are many, but the major ones that you’ll use most often are:
logicalnumeric(integer&double)charactervectormatrixlistdataframe
Understanding the nuanced differences between data types is not important right now. Just know that they exist and that you’ll gain an intuitive understanding of them as you become better aquainted with R. For more information, see the supplemental material on data types and structures.
Packages
User-submitted packages are a huge part of what makes R great. You may hear me use the phrases “base R” during class. What I mean by this is the R that comes as you download it with no packages loaded (sometimes also called “vanilla R”). While it’s powerful in and of itself — you can do everything you need with base R — most of your scripts will make use of one of more contributed packages. These will make your data analytic life much nicer. We’ll lean heavily on the {tidyverse} suite of packages this semester.
Installing packages from CRAN
Many contributed packages are hosted on the CRAN package repository. What’s really nice about CRAN is that packages have to go through quite a few checks in order for CRAN to approve and host them. Checks include making sure the package has documentation, works on a variety of systems, and doesn’t try to do odd things to your computer. The upshot is that you should feel okay downloading these packages from CRAN.
To download a package from CRAN, use:
install.packages("<package name>")
NOTE Throughout this course, if you see something in triangle
brackets (<...>), that means it’s a placeholder for you to change
accordingly.
Many packages rely on other packages to function properly. When you use
install.packages(), the default option is to install all dependencies.
By default, R will check how you installed R and download the right
operating system file type.
Quick exercise
Install the
{tidyverse}package, which is really a suite of packages that we’ll use throughout the semester. Don’t forget to use double quotation marks around the package name.
Loading package libraries
Package libraries can loaded in a number of ways, but the easiest it to write:
library("<library name>")
where "<library name>" is the name of the package/library. You will
need to load these before you can use their functions in your scripts.
Typically, they are placed at the top of the script file.
Quick exercise
Load the
tidyversepackage, which you just installed. This will be a good test of the installation since we will use tidyverse libraries throughout the rest of the workshop.
Help
Even I don’t have every R function and nuance memorized. With all the user-written packages, it would be difficult to keep up if I tried! When stuck, there are a few ways to get help.
Help files
In the console, typing a function name immediately after a question mark will bring up that function’s help file:
## get help file for function
?median
Median Value
Description:
Compute the sample median.
Usage:
median(x, na.rm = FALSE, ...)
Arguments:
x: an object for which a method has been defined, or a numeric
vector containing the values whose median is to be computed.
na.rm: a logical value indicating whether 'NA' values should be
stripped before the computation proceeds.
...: potentially further arguments for methods; not used in the
default method.
Details:
This is a generic function for which methods can be written.
However, the default method makes use of 'is.na', 'sort' and
'mean' from package 'base' all of which are generic, and so the
default method will work for most classes (e.g., '"Date"') for
which a median is a reasonable concept.
Value:
The default method returns a length-one object of the same type as
'x', except when 'x' is logical or integer of even length, when
the result will be double.
If there are no values or if 'na.rm = FALSE' and there are 'NA'
values the result is 'NA' of the same type as 'x' (or more
generally the result of 'x[FALSE][NA]').
References:
Becker, R. A., Chambers, J. M. and Wilks, A. R. (1988) _The New S
Language_. Wadsworth & Brooks/Cole.
See Also:
'quantile' for general quantiles.
Examples:
median(1:4) # = 2.5 [even number]
median(c(1:3, 100, 1000)) # = 3 [odd, robust]
Two question marks will search for the command name in CRAN packages:
## search for function in CRAN
??median
| Package | Topic | Type | Title |
|---|---|---|---|
| bit64 | qtile | help | (Q)uan(Tile)s |
| ellipsis | safe_median | help | Safe version of median |
| future | sticky_globals | help | Place a sticky-globals environment immediately after the global environment |
| ggplot2 | hmisc | help | A selection of summary functions from Hmisc |
| ggpubr | add_summary | help | Add Summary Statistics onto a ggplot. |
| Hmisc | smean.cl.normal | help | Compute Summary Statistics on a Vector |
| httr | guess_media | help | Guess the media type of a path from its extension. |
| httr | parse_media | help | Parse a media type. |
| igraph | time_bins.sir | help | SIR model on graphs |
| matrixStats | rowMedians | help | Calculates the median for each row (column) in a matrix |
| matrixStats | rowWeightedMedians | help | Calculates the weighted medians for each row (column) in a matrix |
| matrixStats | weightedMad | help | Weighted Median Absolute Deviation (MAD) |
| matrixStats | weightedMedian | help | Weighted Median Value |
| olsrr | reference-media | vignette | Media |
| posterior | ess_quantile | help | Effective sample sizes for quantiles |
| posterior | fold_draws | help | Fold draws around their median |
| posterior | mcse_quantile | help | Monte Carlo standard error for quantiles |
| purrr | accumulate | help | Accumulate intermediate results of a vector reduction |
| raster | Summary-methods | help | Summary methods |
| recipes | step_medianimpute | help | Impute Numeric Data Using the Median |
| zoo | rollmean | help | Rolling Means/Maximums/Medians/Sums |
| zoo | zoo | help | Z’s Ordered Observations |
| stats | mad | help | Median Absolute Deviation |
| stats | median | help | Median Value |
| stats | medpolish | help | Median Polish (Robust Twoway Decomposition) of a Matrix |
| stats | runmed | help | Running Medians - Robust Scatter Plot Smoothing |
| stats | smooth | help | Tukey’s (Running Median) Smoothing |
| stats | smoothEnds | help | End Points Smoothing (for Running Medians) |
| survival | Math.Surv | help | Methods for Surv objects |
At first, using help files may feel like trying to use a dictionary to see how to spell a word — if you knew how to spell it, you wouldn’t need the dictionary! Similarly, if you knew what you needed, you wouldn’t need the help file. But over time, they will become more useful, particularly when you want to figure out an obscure option that will give you exactly what you need.
Google it!
Google is a coder’s best friend. If you are having a problem, odds are a 1,000+ other people have too and at least one of them has been brave enough to ask about it in a forum like StackOverflow, CrossValidated, or R-help mailing list.
If you are lucky, you’ll find the exact answer to your question. More likely, you’ll find a partial answer that you’ll need to modify for your needs. Sometimes, you’ll find multiple partial answers that, in combination, help you figure out a solution. It can feel overwhelming at first, particularly if it’s a way of problem-solving that’s different from what you’re used to. But it does become easier with practice.
Google it!
Asking for help: order of operations
When needing help for this class, your order of operations should be:
- Try a lot on your own
- R help files
- Class peer
- Me
This is not because I don’t want to help. My concern is the opposite: that I’m likely to just show you. Data analysis is tricky because no two problems are alike. But over time, they do rhyme. The time you put in now learning to figure things out on your own will be well paid in the future.

